In biology textbooks and beyond, the human genome and DNA therein typically are taught in only one dimension. While it can be helpful for learners to begin with the linear presentation of how stretches of DNA form genes, this oversimplification undersells the significance of the genome’s 3D structure.
To fit in the nucleus of our cells, six feet of DNA is wound up like thread on protein spools called histones. In its packaged form called chromatin, coiled up DNA features many loops and clumps. While it may look random and messy to the untrained eye, these tumbleweed-like shapes bring certain genomic regions into close contact while sheltering others.
Problems with this 3D structure are associated with many diseases including developmental disorders and cancer. Almost 12% of genomic regions in breast cancer cells have incurred issues with their chromatin structure, while other structural issues are known to cause T-cell acute lymphoblastic leukemia.
Scientists at Sanford Burnham Prebys and colleagues in Hong Kong published findings June 27, 2025, in Genome Biology demonstrating a new approach for better understanding chromatin’s 3D structure and its influence.
The research team hypothesized that the 3D shape of regions of the genome influences how genes are regulated.
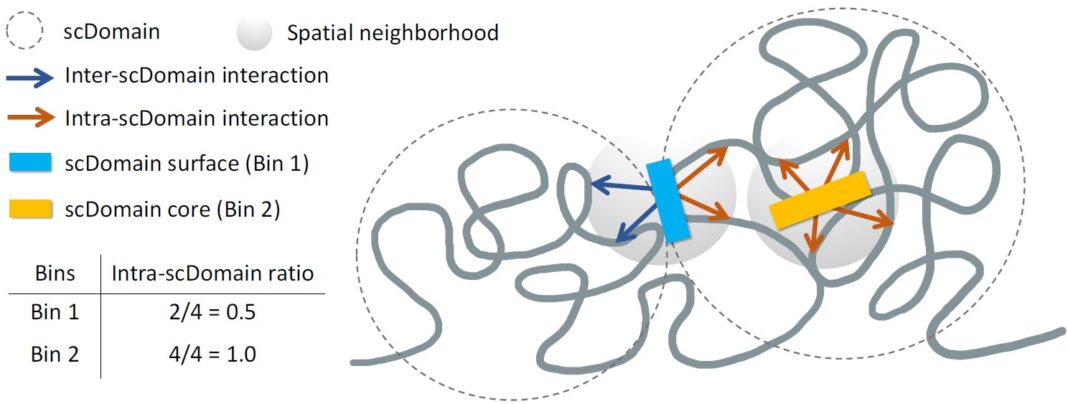
“We know that many regions of the genome tend to form what are known as topologically associating domains or TADs,” said Kelly Yichen Li, Ph.D., a postdoctoral associate at Sanford Burnham Prebys and lead author of the study. “Parts of the genome within these domains can interact more frequently with each other, while they tend to be isolated from the region outside this domain.”
The investigators noticed when taking many images of chromatin to conduct spatial mapping experiments, TAD-like regions of the genome in individual cells tended to take a globular shape, albeit with the varied bumpiness and spherical irregularity of a supermarket’s selection of potatoes. Certain characteristics of these regions in the 3D images suggested they may influence the function of nearby genes.
“If you picture these clumps of chromatin fiber being roughly in the shape of a potato, we predicted that regions of the genome closer to the surface are more active due to exposure to nearby biochemical signals in the cell nucleus,” said Yuk-Lap (Kevin) Yip, Ph.D., a professor and the interim director of the Center for Data Sciences at Sanford Burnham Prebys, and the senior and corresponding author of the manuscript.
Similar to the protection offered by a potato’s fibrous skin to its starchy flesh, the scientists predicted that signals promoting gene expression would have more difficulty reaching regions of the genome buried near the core of a globular wad of chromatin. To test this, they developed a method of measuring a genomic region’s proximity to the isolated center of a chromatin clump.
“We used a metric to quantify the ‘coreness’ of a genomic region in a chromatin domain,” said Li. “This measure also allowed us to define the surface and core, and we went on to show that surface regions are more active than core regions.”
“The type of data we can apply this measure to is becoming quite plentiful,” said Yip. “There is a lot of potential to study how coreness links to gene activity and disease in different cell types.”
Yip and Li plan to continue collaborating with the lab of Pier Lorenzo Puri, MD, to advance our understanding of how the 3D structure of the genome affects muscle stem cell development and the progression of muscular dystrophy.
More information:
Kelly Yichen Li et al, Regulatory roles of three-dimensional structures of chromatin domains, Genome Biology (2025). DOI: 10.1186/s13059-025-03659-7
Provided by
Sanford-Burnham Prebys
Citation:
Cutting to the core of how 3D structure shapes gene activity (2025, July 11)
retrieved 12 July 2025
from https://phys.org/news/2025-07-core-3d-gene.html
This document is subject to copyright. Apart from any fair dealing for the purpose of private study or research, no
part may be reproduced without the written permission. The content is provided for information purposes only.